[摘要] 背景与目的:结直肠癌(colorectal cancer,CRC)是危害全球人类生命健康的主要恶性肿瘤之一,其发病率和死亡率长期居高不下。钾离子通道调节因子1(potassium channel modulatory factor 1,KCMF1)属于E3泛素连接酶家族的一员,通过RING结构域与靶蛋白结合,参与调节体内多种生物学过程。然而,KCMF1在CRC中的作用尚不清楚。本研究旨在探索KCMF1在CRC中的表达情况,并探讨其对CRC细胞增殖的影响及可能的分子机制。方法:利用癌症基因组图谱(The Cancer Genome Atlas,TCGA)和基因型-组织表达(Genotype-Tissue Expression,GTEx)数据库,分析CRC组织中KCMF1的表达水平及其与CRC患者预后的相关性。通过免疫组织化学法(immunohistochemistry,IHC)检测90例配对的人CRC组织样本中KCMF1的蛋白表达水平。通过慢病毒感染肠癌HCT116和HCT15细胞,转导针对KCMF1基因的短发夹RNA(shKCMF1),分别采用四甲基偶氮唑盐(methyl thiazoyl terazolium,MTT)实验和克隆形成实验检测敲降KCMF1对细胞增殖的影响;采用蛋白质印迹法(Western blot)和流式细胞学实验检测敲降KCMF1对细胞凋亡和细胞周期的影响;利用转录组测序(RNA sequencing,RNA-Seq)检测敲降KCMF1对HCT116细胞的转录谱的影响,运用生物信息学分析受KCMF1调控的信号通路;利用实时荧光定量聚合酶链反应(real-time fluorescence quantitative polymerase chain reaction,RTFQ-PCR)、Western blot、荧光素酶报告基因实验及细胞免疫荧光实验等验证相关信号通路的改变。结果:TCGA和GTEx数据库分析以及IHC结果显示,与癌旁组织相比,CRC组织中KCMF1 mRNA的表达及蛋白水平均显著升高(P<0.01),其表达水平与患者的生存时间呈负相关(P<0.01),并与CRC临床分期呈正相关(P<0.05)。与对照细胞相比,敲降KCMF1的HCT116和HCT15细胞的增殖能力显著降低(P<0.001),细胞凋亡水平显著增高(P<0.001),细胞周期停滞在G1期(P<0.01)。RNA-Seq分析发现,KCMF1参与调控核因子-κB(nuclear factor-κB,NF-κB)等多个信号通路。敲降KCMF1后,NF-κB信号通路下游靶基因BCL-XL、XIAP和CIAP的转录水平降低(P<0.05),p65的磷酸化水平下降,同时p65的核转移受抑制(P<0.01),NF-κB信号报告基因活性降低(P<0.01)。结论:KCMF1在人CRC组织中呈高表达,并与患者的高临床分期和不良预后呈正相关;KCMF1可能通过激活NF-κB信号通路促进CRC细胞增殖。KCMF1可能是CRC的一个潜在治疗新靶点。
[关键词] 结直肠癌;钾离子通道调节因子1;细胞凋亡;核因子-κB信号通路
[Abstract] Background and purpose: Colorectal cancer (CRC) is one of the major malignant tumors threatening human health worldwide, with long-term high incidence and mortality rate. Potassium channel modulatory factor 1 (KCMF1) is a member of the E3 ubiquitin ligase family. It binds to target proteins through the RING domain and participates in the regulation of a variety of biological processes in vivo. However, the function of KCMF1 in CRC remains unclear. This study aimed to investigate the expression level of E3 ubiquitin ligase KCMF1 in colorectal tumor, and to explore the effects of KCMF1 on the proliferation of CRC cells and its underlying molecular mechanism. Methods: The The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases were used to analyze the expression level of KCMF1 in CRC tissues and adjacent tissues and the association between the KCMF1 expression and the prognosis of CRC patients. Furthermore, immunohistochemical staining was performed to detect the protein level of KCMF1 in 90 paired human CRC tissues and adjacent non-tumor tissues. Lentiviral shRNA delivery system was employed to specifically target the KCMF1 gene (shKCMF1) in HCT116 and HCT15 CRC cell lines. The effects of KCMF1 knockdown on cell proliferation, apoptosis and cell cycle distribution were assessed by methyl thiazoyl terazolium (MTT) assay, colony formation assay, Western blot and flow cytometry. Changes in the transcriptional profile in HCT116 cells upon KCMF1 knockdown were identified by RNA sequencing (RNA-Seq), and the affected signaling pathways were evaluated by bioinformatics analysis. Real-time fluorescence quantitative polymerase chain reaction (RTFQ-PCR), Western blot, luciferase reporter assay and cell immunofluorescence assay were utilized to validate the alteration of the affected signaling pathway. Results: The TCGA and GTEx databases and IHC results showed that the mRNA and protein expression levels of KCMF1 in CRC tissues were significantly upregulated compared with adjacent tissues (P<0.01). KCMF1 expression level was negatively correlated with the survival time of patients with CRC (P<0.01), and was positively associated with CRC clinical stage (P<0.05). Compared with control cells, KCMF1 knockdown significantly inhibited the proliferation of HCT116 and HCT15 cells (P<0.001), induced cell apoptosis (P<0.001), and led to cell cycle arrest in G1 phase (P<0.01). RNA-Seq analysis showed that KCMF1 was involved in the regulation of several signaling pathways, including nuclear factor-κB (NF-κB) signaling pathway. KCMF1 knockdown reduced the transcription levels of the target genes of NF-κB signaling pathway, including BCL-XL, XIAP and CIAP (P<0.05), and suppressed the expression of phosphorylated p65 and nuclear translocation of p65 (P<0.01). Meanwhile, the activity of NF-κB reporter was reduced in tumor cells upon KCMF1 knockdown (P<0.01). Conclusion: The expression of KCMF1 is significantly upregulated in human CRC tissues and positively associated with advanced clinical stage and poor prognosis. KCMF1 may promote the proliferation of CRC cells by activating the NF-κB signaling pathway. KCMF1 may be a potential new therapeutic target for CRC.
[Key words] Colorectal cancer; Potassium channel modulatory factor 1; Apoptosis; Nuclear factor-κB signaling pathway
结直肠癌(colorectal cancer,CRC)是临床常见的消化系统恶性肿瘤,发病率在所有癌症类型中位居第3位,死亡率位居第2位[1]。在其恶性转化过程中,通常伴有众多的分子突变或多个信号通路的异常调控[2-3]。尽管目前CRC患者的诊治效果有所改善,但对于晚期及发生转移的患者,仍存在诸多不足,如治疗反应率低、不良反应大、治疗抵抗等[4-7]。因此,亟待对促进CRC发生、发展的靶基因及其分子机制进行深入研究。CRC的发生、发展与泛素-蛋白酶体系统密切相关。E3泛素连接酶是决定泛素-蛋白酶体系统特异性的重要组分,通过介导靶蛋白的识别和泛素化修饰,参与调控细胞信号转导、细胞凋亡、细胞周期及DNA损伤修复等生物学过程[8-9]。在CRC的发生、发展过程中,E3泛素连接酶的功能经常发生异常,导致原癌蛋白转化为癌蛋白、致癌病毒复制或相关肿瘤抑制因子失活等,促进肿瘤的转化和形成[10]。
钾离子通道调节因子1(potassium channel modulatory factor 1,KCMF1)属于E3泛素连接酶家族的一员,是一种进化上高度保守的锌指蛋白[11]。KCMF1与RAD6及UBR4参与构成泛素连接酶E2-E3复合体,对蛋白的泛素编码进行重编程,使其发生溶酶体介导的自噬和降解[12]。在应激条件下,KCMF1通过泛素化途径帮助调节错误折叠蛋白聚集体和细胞器的周转[13-14]。在多种肿瘤类型中,KCMF1表达增加并促进肿瘤细胞的恶性生物学行为[15-16]。然而,目前KCMF1在CRC中的作用及其分子机制仍不清楚,有待进一步探索和研究。
本研究旨在通过癌症基因组图谱(The Cancer Genome Atlas,TCGA)、基因型-组织表达(Genotype-Tissue Expression,GTEx)数据库及临床组织芯片,应用生物信息学方法分析KCMF1在CRC中的表达水平及其与患者生存预后的相关性。利用细胞学实验揭示其对CRC细胞增殖的影响及其相关机制。通过基因集富集分析(gene set enrichment analysis,GSEA)探讨KCMF1可能参与调节的信号通路,探索其调节CRC发生、发展的具体分子机制,为CRC的预后和治疗提供新的思路。
1 材料和方法
1.1 组织样本来源及临床资料
CRC组织微阵列(HCol-Ade180Sur-01,90例)购自上海芯超生物科技有限公司。手术时间为2006年7月—2007年5月,随访时间为2006年11月—2013年8月,其中男性47例,女性43例;≤65岁34例,>65岁56例;T1期7例,T2期47例,T3期32例,T4期2例,2例分期不明;生存期为3~85个月,平均生存期51个月,7例无随访记录。所有程序均在共识协议下进行,并通过上海芯超生物科技有限公司伦理委员会的审批(伦理批号:SHYJS-CP-1404014)。
1.2 细胞、试剂和仪器
人CRC细胞系HCT116、HCT15以及人胚肾细胞HEK-293T细胞均由上海交通大学医学院附属仁济医院实验室保存。细胞培养基DMEM购自上海源培生物科技股份有限公司,胎牛血清(fetal bovine serum,FBS)购自美国Gibco公司,四甲基偶氮唑盐(methyl thiazoyl terazolium,MTT)、碘化丙啶(propidium iodide,PI)、7-氨基放线菌素D(7-aminoactinomycin D,7-AAD)和嘌呤霉素(puromycin,puro)均购自美国Sigma-Aldrich公司,质粒小抽提试剂盒和胶回收试剂盒购自天根生化科技(北京)有限公司,LipofectamineTM2000、optiDMEM和总RNA提取试剂(total RNA extractor,TRIzol)均购自美国Invitrogen公司,限制性核酸内切酶购自美国New England Biolabs公司,T4 DNA连接酶购自上海昂一生物科技有限公司,高保真酶DNA聚合酶、RNA反转试剂盒和以SYBR Green为染料用于实时荧光定量聚合酶链反应(real-time fluorescence quantitative polymerase chain reaction,RTFQ-PCR)的SYBR Premix Ex TaqTM试剂盒均购自上海翊圣生物科技有限公司。兔抗人KCMF1单克隆抗体购自美国Invitrogen公司,兔抗人p65单克隆抗体、兔抗人p-p65单克隆抗体、兔抗人p-IKK单克隆抗体、兔抗人IKK单克隆抗体、兔抗人p-IκB单克隆抗体和兔抗人IκB单克隆抗体均购自美国Cell signaling technology公司,鼠抗人GAPDH单克隆抗体购自武汉爱博泰克生物科技有限公司,Western blot一抗稀释液购自上海碧云天生物技术股份有限公司,免疫染色试剂盒购自美国Gene Tech公司。
pLVX-IRES-puro-cflag、pLKO.1-puro表达质粒及阴性对照质粒由上海交通大学医学院附属仁济医院实验室提供。KCMF1基因的上游引物序列为5’-CCGCTCGAGGCCACCATGTCCC GACATGAAGGTG-3’,下游引物序列为5’-GCT CTAGAAAGAGGAGGTGGTGGAGGCTCATTT CC-3’。特异性针对KCMF1基因的shRNA序列1为5’-CGTAGATCAAACATGCACTTT-3’,序列2为5’-GCAGTTCTACTGGTGGACTTT-3’,引物均由上海擎科生物科技有限公司合成。
ViiA 7 RTFQ-PCR仪、生物安全柜和CO2细胞培养箱均购自美国Thermo Fisher Scientific公司,倒置荧光显微镜购自日本Olympus公司,Odyssey双色红外激光成像仪购自美国LI-COR公司,多功能酶标仪购自美国BioTek公司。
1.3 实验方法
1.3.1 细胞培养
人CRC细胞及HEK-293T使用含10% FBS和1%的青霉素-链霉素的完全DMEM高糖培养基培养,细胞培养箱内环境设置为37 ℃、CO2体积分数为5%,当细胞融合度达80%时进行传代。
1.3.2 病毒包装与感染
采用聚乙烯亚胺(polyethylenimine,PEI)转染HEK-293T细胞。HEK-293T细胞于37 ℃、CO2体积分数为5%的培养箱中培养,至细胞融合度达30%~40%。按目的质粒∶包装质粒(psPAX2)∶包膜质粒(pMD2.G)=3∶2∶1的比例计算各质粒的用量,共转染到HEK-293T细胞中,6 h后更换培养基,分别于48和72 h收集病毒液。
待HCT116和HCT15细胞融合度为50%时,将病毒液与聚凝胺混匀后加入细胞中,24 h后重复感染1次,48 h后更换培养基,并用puro进行筛选,得到稳定转入目的质粒的细胞。
1.3.3 MTT实验
在96孔板中接种HCT116和HCT15细胞,分为shCon组(对照组)、shKCMF1-1组和shKCMF1-2组,每组3个复孔,每孔接种1 000个细胞,置于恒温细胞培养箱中培养。第2天,往待测孔中加入20 µL 5 mg/mL的MTT溶液,继续温育3~4 h,弃去培养液,加入200 µL 二甲基亚砜(dimethyl sulfoxide,DMSO)溶液重悬,用酶标仪检测490 nm处各孔细胞的吸光度(D)值。
1.3.4 克隆形成实验
将消化转染后的HCT116和HCT15细胞(分组同1.3.3节)接种于6孔板中,于培养箱中培养至肉眼可见集落形成,弃去培养基,采用甲醇固定细胞20 min,结晶紫染色过夜,冲洗晾干后采集图像。
1.3.5 流式细胞凋亡检测
在12孔板中接种HCT116和HCT15细胞(分组同1.3.3节),在恒温培养箱中温育72 h,取培养上清液及消化收集的细胞于流式管中。用预冷的磷酸缓冲盐溶液(phosphate-buffered saline,PBS)清洗重悬,加1 mL 1×结合缓冲液。细胞染色及分析:加入20 µL染色工作液(含2 µL APC-Annexin Ⅴ和0.1 µL 7-AAD),4 ℃避光温育25 min,加入200 µL 1×结合缓冲液稀释,用BD流式细胞仪按照标准程序进行检测。
1.3.6 细胞周期检测
在6孔板中接种HCT116和HCT15细胞(分组同1.3.3节),在恒温培养箱中温育48 h,更换无血清培养液,24 h后更换含10% FBS的细胞培养基,20 h后收集细胞,加1 mL的75%乙醇溶液固定细胞,第2天采用PI染色,4 ℃避光温育 30 min,加入300 µL PBS稀释,用BD流式细胞仪按照标准程序进行检测。
1.3.7 RTFQ-PCR
收集HCT116和HCT15细胞(分组同1.3.3节),用TRIzol试剂提取细胞总RNA,反转录为cDNA。采用SYBR法,在ViiA 7 RTFQ-PCR仪上进行扩增。BCL-XL基因的上游引物序列为5’-CGGATTTGAATCTCTTTCTCTCCC-3’,下游引物序列为5’-CGACCCCAGTTTAC CCCATC-3’;CIAP基因的上游引物序列为5’-GGCCGTATCTCCTTGTCGG-3’,下游引物序列为5’-TCAGGGTTGTAAATCGCAGTG-3’;XIAP基因的上游引物序列为5’-ACTTCGGG TTTCACGACTCC-3’,下游引物序列为5’-CTTG TCCACCTTTTCGCGCC-3’;内参照GAPDH基因的上游引物序列为5’-CATGAGAAG TATGACAACAGCCT-3’,下游引物序列为5’-AGTCCTTCCACGATACCAAAGT-3 ’;KCMF1基因的上游引物序列为5’-TGGA GGCGATCCTAATCATG-3’,下游引物序列为5’-CTCAGTTATCAGGAGTGAGA-3’。引物均由上海擎科生物科技有限公司合成。采用2-ΔΔCt法比较目的基因的相对表达量。
1.3.8 蛋白质印迹法(Western blot)
收集HCT116和HCT15细胞(分组同1.3.3节),裂解细胞并提取细胞总蛋白,用二辛可宁酸(bicinchoninic acid,BCA)法定量,经10%十二烷基硫酸钠聚丙烯酰胺凝胶电泳(sodium dodecylsulphate polyacrylamide gel electrophoresis,SDS-PAGE)分离蛋白,转移至硝酸纤维素膜上,用含5%牛血清白蛋白的封闭液封闭60 min,加入相应一抗,置于4 ℃摇床过夜。第2天用含有吐温-20三乙醇胺缓冲盐溶液(tris-buffered saline Tween,TBST)洗涤,加入同源二抗,室温避光2 h,TBST洗涤,由Odyssey Sa双色成像系统获取图像。
1.3.9 免疫组织化学法(immunohistochemistry,IHC)
采用IHC检测90例CRC患者肿瘤组织及对应的癌旁组织中KCMF1的表达情况。石蜡包埋组织切片脱蜡、水化、抗原修复、封闭后,加入一抗[兔抗人KCMF1单克隆抗体(体积稀释比例为1∶200)],4 ℃温育过夜。第2天滴加二抗,采用3,3’-二氨基联苯胺(3,3’-diaminobenzidine,DAB)显色剂显色,苏木精复染,脱水后中性树脂封存。IHC评分H-score=Σpi(i+1),其中i代表染色强度评分(阴性为0分,弱阳性为1分,中等阳性为2分,强阳性为3分),pi代表染色面积评分(pi表示阳性染色细胞数量占切片中所有细胞数量的百分数,分别记录各个染色强度细胞所占百分数),最终评分区间为0~300。
1.3.10 转录组测序(RNA sequencing,RNA-Seq)及生物信息学分析
在HCT116细胞中敲降KCMF1基因,扩大培养后,经过puro筛选3 d后,用TRIzol提取总RNA送至上海擎科生物科技有限公司进行测序,测序平台为Illumina NovaSeq,建库时采取Illumina TruseqTM RNA sample prep Kit方法。测序后采用RSEM软件对基因表达进行定量分析,采用序列数量计算基因表达水平。差异表达基因分析采用DESeq2软件,差异定义标准为FDR<0.05及 |log2 FC|≥1,然后对差异表达的基因进行基因本体论(gene ontology,GO)分析、GSEA等生物信息学处理。
1.3.11 统计学处理
使用GraphPad Prism 9.0软件对实验数据进行统计分析并作图,计量资料以x±s表示。两组间的数据分析采用Student t检验,3组间及以上的数据分析采用单因素方差分析,在发现显著差异后,使用Student-Newman-Keuls方法进行事后检验以确定存在显著差异的组别。生存分析采用Kaplan-Meier绘制曲线,采用log-rank检验比较组间预后差异。针对IHC实验结果比较CRC组织与癌旁组织中的KCMF1蛋白表达差异采用独立样本t检验。KCMF1表达差异与临床TNM分期的关联性采用χ2检验。相关性分析采用Spearman秩相关检验。差异基因分析结果中P值为显著性检验的P值,计算模型为负二项分布。在HALLMARK gene signatures富集分析中P值为显著性检验的P值,其计算基于超几何分布原理。P<0.05为差异有统计学意义。
2 结 果
2.1 人CRC组织细胞KCMF1的表达水平及其与患者预后的相关性
首先利用TCGA和GTEx数据库对KCMF1进行泛癌分析,研究其在肿瘤组织和癌旁组织中的表达差异。结果显示,KCMF1在结肠癌和直肠癌组织中表达均明显增加(P<0.001,图1A)。对GSE39582数据集临床队列进行Kaplan-Meier曲线生存分析,结果显示,CRC组织中KCMF1的表达水平与患者的不良预后呈正相关(P<0.01,图1B)。为验证上述结果,通过IHC分析90例临床上配对的人CRC组织和癌旁组织中KCMF1蛋白的水平。结果显示,CRC组织中KCMF1的染色强度明显高于癌旁组织 (P<0.01,图1C、1D)。此外,根据患者的生存期信息进行统计分析,结果显示,KCMF1高表达组患者生存期明显缩短(P<0.01),并与较高的TNM分期呈正相关(P<0.05,图1E、 1F)。上述结果提示KCMF1在结直肠肿瘤的发生、发展过程中发挥着重要作用。
2.2 敲降KCMF1抑制CRC细胞体外增殖
为探究KCMF1在CRC生物学表型方面的调节作用,本研究首先构建了稳定敲降KCMF1的CRC细胞系HCT116和HCT15。Western blot结果显示,与shCon组相比,shKCMF1组细胞中KCMF1的蛋白表达水平明显下调(图2A)。MTT及克隆形成实验结果显示,与对照组相比,KCMF1敲降组中CRC细胞的增殖能力明显受到抑制(P<0.001,图2B、2C)。进一步采用Western blot检测敲降KCMF1对CRC细胞中凋亡相关蛋白表达水平的影响,结果显示,敲降KCMF1后,促凋亡分子BAX蛋白的表达水平升高,而抗凋亡分子BCL-2和BCL-XL蛋白的表达水平降低(图2D);流式细胞术分析结果同样显示,KCMF1敲降组的细胞凋亡比例显著增加,而加入pan-caspase抑制剂Z-VAD处理能够逆转KCMF1缺失诱导的细胞凋亡(P<0.001,图2E)。此外,采用Western blot检测敲降KCMF1对CRC细胞中细胞周期蛋白表达水平的影响,结果显示,KCMF1敲降后,细胞周期蛋白D1和细胞周期蛋白依赖性激酶4(cyclin dependent kinase 4,CDK4)的蛋白表达水平降低(图2F);流式细胞术分析结果显示,敲降KCMF1会导致细胞周期停滞在G1期(P<0.01,图2G)。综上所述,KCMF1表达下调能够抑制CRC细胞的体外增殖,导致细胞凋亡及细胞周期停滞在G1期。
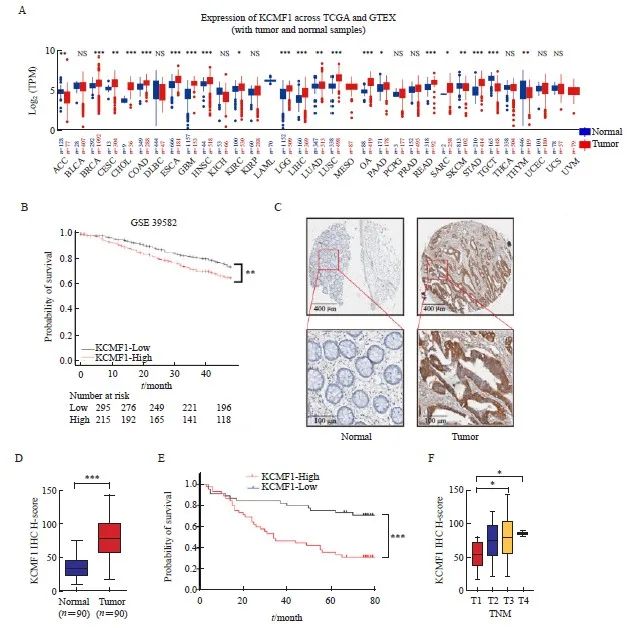
图1 CRC组织中KCMF1高表达且与患者不良预后相关
Fig. 1 High expression of KCMF1 is associated with poor prognosis of patients with CRC
A: mRNA levels of KCMF1 in different types of tumor and normal tissues from the TCGA and GTEx databases. TPM: Transcripts per million; NS: No significance; *: P<0.05, assessed by Student’s t-tests; **: P<0.01, assessed by Student’s t-tests; ***: P<0.001, assessed by Student’s t-tests. B: The correlation between KCMF1 expression and the prognosis of CRC patients in GSE39582 dataset from the GEO database. **: P<0.01, assessed by log-rank tests. C: Representative images of immunohistochemical staining for KCMF1 in CRC tissues and their paired adjacent tissues. D: Immunohistochemical staining scores in the 90 colorectal tumor tissues and their paired adjacent tissues. ***: P<0.001, assessed by Student’s t-tests. E: The survival probability of 90 patients with high or low expression of KCMF1 in colorectal tumors; The KCMF1 scores are ranked by High and Low, with KCMF1-High group (n=45) and KCMF1-Low group (n=45). ***: P<0.001, assessed by log-rank tests. F: The correlation between KCMF1 expression and TNM stage in patients with CRC. *: P<0.05, assessed by Student-Newman-Keuls.
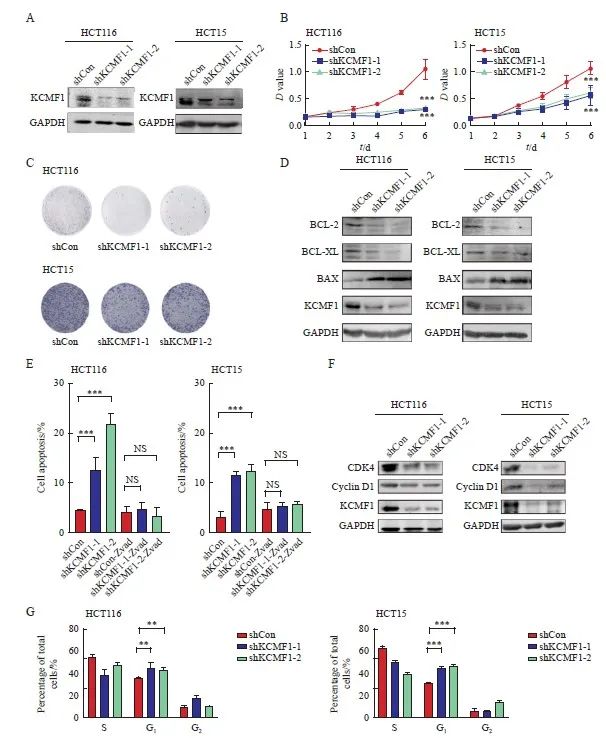
图2 敲降KCMF1对结肠癌细胞体外增殖的调节作用
Fig. 2 Effects of KCMF1 knockdown on the proliferation of CRC cells in vitro
A: Expression level of KCMF1 protein in HCT116 and HCT15 cells with or without shKCMF1 expression. B, C: The proliferation of HCT116 and HCT15 cells with or without shKCMF1 expression was assessed by MTT assay (B) and colony formation assay (C). ***: P<0.001, assessed by Student’s t-tests. D: Expression levels of the apoptosis-related proteins in HCT116 and HCT15 cells with or without shKCMF1. E: Flow cytometry analysis of the percentage of apoptotic cells in the indicated cells treated with or without Z-VAD-FMK (20 µm) for 48 h. NS: No significance; ***; P<0.001, assessed by Student-Newman-Keuls. F: Expression levels of the cell cycle-related proteins in HCT116 and HCT15 cells with or without shKCMF1. G: Flow cytometry analysis of the cell cycle distribution of HCT116 and HCT15 cells with or without shKCMF1. **: P<0.01, assessed by Student-Newman-Keuls; ***: P<0.001, assessed by Student-Newman-Keuls.
2.3 敲降KCMF1对CRC细胞信号通路的影响
为研究KCMF1调控CRC细胞增殖的可能分子机制,本研究对KCMF1敲降和对照HCT116细胞进行RNA-Seq,对筛选出的差异表达基因进行GO富集分析。结果显示,KCMF1敲降影响了细胞凋亡、细胞周期等生物学过程相关基因的表达(图3A、3B)。利用HALLMARK特征信号集和GSEA分别进行富集分析,结果显示,肿瘤细胞中多个信号通路可被KCMF1调控;其中,HALLMARK_TNFA_SIGNALING_VIA_NFKB通路被富集(图3C、3D),提示KCMF1可能参与调节核因子-κB(nuclear factor-κB,NF-κB)信号通路。利用TCGA数据库分析KCMF1与NF-κB特征信号集表达水平的关系,结果显示,两者呈显著正相关(P<0.001,图3E)。在TCGA数据集中,KCMF1高表达伴NF-κB信号通路活化的患者预后最差,KCMF1低表达伴NF-κB信号通路低活性的患者预后较好(P<0.01,图3F)。
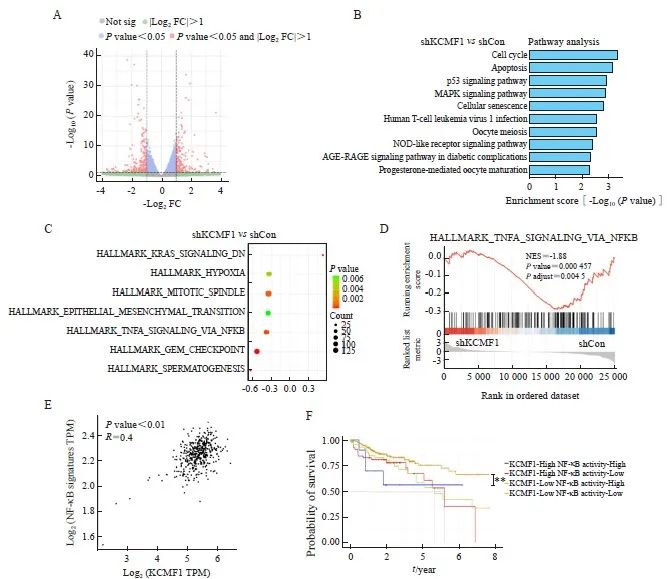
图3 敲降KCMF1对CRC细胞信号通路的影响
Fig. 3 Effects of KCMF1 knockdown on the signaling pathways in human CRC cells
A: Volcano plots show differentially expressed genes affected by KCMF1 knockdown in HCT116 cells. The significant different genes are determined with the following criteria: |Log2 FC|>1 and P<0.05. FC: Fold change. B: GO analysis of the down-regulated genes in HCT116 cells with KCMF1 knockdown. P<0.05 and Log2 FC≤-1 were used as the criteria to define down-regulated genes. C: Enrichment analysis of the KCMF1-regulated genes with HALLMARK gene signatures, the significantly enriched signatures were ranked by P value, the bar plot shows the top 10 signatures. D: GSEA of the transcriptional profiles of KCMF1-koncokdown HCT116 cells and control cells with the TNFα-NF-κB signature. E: Spearman correlation analysis was performed between BIOCARTA_NFKB_PATHWAY and KCMF1 expression based on TCGA database cohort. F: The TCGA cohort were classified into four groups by both KCMF1 expression and NF-κB activity and subjected to Kaplan–Meier overall survival analysis. The activity of NF-κB signaling was determined by the GSVA score of the gene set BIOCARTA_NFKB_SIGNALING. **: P<0.01, assessed by log-rank tests.
2.4 敲降KCMF1抑制CRC细胞中NF-κB信号的活化
为进一步验证转录组学的结果,本研究在稳定敲降KCMF1的CRC细胞中检测NF-κB信号通路的活化情况。RTFQ-PCR结果显示,在KCMF1敲降组中,NF-κB信号通路下游靶基因BCL-XL、XIAP和CIAP的mRNA水平均降低(P<0.05,图4A)。Western blot结果显示,敲降KCMF1后,p65、IKK和IκB的磷酸化水平受到明显抑制(图4B)。荧光素酶报告基因实验结果显示,KCMF1缺失能够抑制NF-κB信号通路的活性(P<0.01,图4C)。此外,Western blot及细胞免疫荧光分析结果显示,敲降KCMF1能够抑制NF-κB蛋白亚基p65向核内的转移(P<0.01,图4D~4F)。综上所述,敲降KCMF1能够抑制CRC细胞中NF-κB信号通路的活化。
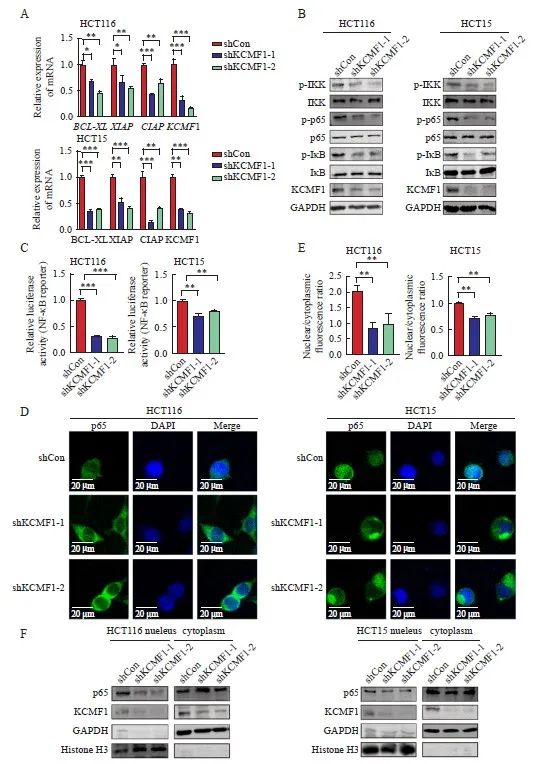
图4 敲降KCMF1抑制人CRC细胞中NF-κB信号通路的活化
Fig. 4 Knockdown of KCMF1 inhibits the activation of the NF-κB signaling in human CRC cells
A: mRNA levels of KCMF1, BCL-XL, XIAP and CIAP in HCT116 and HCT15 cells with or without KCMF1 knockdown. *: P<0.05, assessed by Student-Newman-Keuls; **: P<0.01, assessed by Student-Newman-Keuls; ***: P<0.001, assessed by Student-Newman-Keuls. B: Expression of the phosphorylated proteins related to NF-κB signaling in HCT116 and HCT15 cells with or without KCMF1 knockdown. C: Activity of the NF-κB-reporter in HCT116 and HCT15 cells with KCMF1 knockdown. **: P<0.01, assessed by Student-Newman-Keuls; ***: P<0.001, assessed by Student-Newman-Keuls. D: Immunofluorescence of p65 (green) in HCT116 and HCT15 cells treated with TNFα (10 ng/mL) for 5 min after KCMF1 knockdown. DAPI stains nuclear DNA (blue). Scale=20 microns. E: Quantification of p65 nuclear and cytosolic localization was assessed in the indicated cells, as shown by a ratio of nuclear to cytoplasmic fluorescence. **: P<0.01, assessed by Student-Newman-Keuls. F: p65 cellular localization in HCT116 and HCT15 cells with or without KCMF1 knockdown.
3 讨 论
CRC是一种恶性程度很高的消化系统恶性肿瘤,目前发病年龄正趋向于年轻化[17]。尽管针对CRC已经开展了大量研究,治疗手段也有了较大进步,但晚期及发生转移的患者仍存在治疗反应率低且易复发等问题,导致其预后较差[2,6,18],急需寻找新的可靠治疗靶点。
KCMF1是属于RING finger家族的E3泛素连接酶,参与蛋白质泛素化修饰过程[12]。多项研究[12-13,19]表明,KCMF1通过RING结构域与靶蛋白结合,介导其自噬或降解,从而参与调节体内多种生物学过程。以往研究[11,15-16]表明, KCMF1与人类多种癌症的发生、发展有关,能够调控肿瘤细胞的增殖、侵袭、转移及凋亡等过程。在胰腺癌中,KCMF1能够增强肿瘤细胞的增殖、迁移和侵袭能力,并抑制细胞周期蛋白D1和CDK4的蛋白表达水平,延缓小鼠胰腺肿瘤的形成[11]。另外,敲降KCMF1可抑制结肠癌干细胞的增殖和细胞集落的形成[20]。上述研究结果提示KCMF1在多种类型的肿瘤组织中高表达并与肿瘤的发生、发展密切相关,具有促癌作用。因此,KCMF1可能是多种人类癌症的潜在生物标志物。但KCMF1在人类CRC中的作用和具体分子机制尚缺乏深入的研究。
本研究通过对TCGA、GTEx数据库以及临床组织芯片中的信息进行分析,发现CRC组织中KCMF1的表达水平显著增加,并且KCMF1高表达与患者的不良预后及临床分期相关,预示KCMF1可能作为新的CRC辅助诊断的标志物。本研究通过体外实验进一步探讨KCMF1在CRC中的具体作用。结果显示,敲降KCMF1可以抑制CRC细胞增殖,促进CRC细胞凋亡,并诱导细胞周期发生阻滞,表明KCMF1在CRC的发生、发展过程中发挥重要作用。
为探讨KCMF1缺失抑制CRC细胞增殖的分子机制,本研究进行了RNA-Seq分析。结果显示,敲降KCMF1能够影响NF-κB信号通路相关基因的表达。NF-κB信号通路参与调控CRC细胞的增殖和凋亡。在CRC中,NF-κB信号通路呈组成性激活状态,抑制NF-κB的激活可以诱导CRC细胞发生凋亡[21]。UBS109通过降低NF-κB的活化下调E2F转录因子1的表达,从而抑制细胞周期进程,使CRC细胞HCT116和HT-29被阻滞在G0/G1期边界,导致肿瘤生长速度显著降低[22]。另外,阿昔平可以抑制CRC细胞系中NF-κB的激活,使c-FLIP和MCL-1的表达降低,并增强细胞对肿瘤坏死因子相关凋亡诱导配体诱导的细胞死亡敏感性,刺激细胞凋亡[23]。
本研究进一步分析了敲降KCMF1对CRC细胞NF-κB信号相关分子的调节作用。结果显示,KCMF1敲降能够下调p65、IKK和IκB的磷酸化水平,抑制NF-κB信号下游靶基因的转录水平,并减少p65的核转移。这些数据以及上述转录谱分析结果表明,KCMF1具有促进调控NF-κB信号通路的作用。鉴于NF-κB信号通路对结直肠细胞增殖的促进作用,我们推测KCMF1可能通过调控NF-κB信号通路进而影响CRC细胞的增殖。既往研究表明,E3泛素连接酶可以通过泛素化修饰方式调控NF-κB信号通路的转录及激活[24]。过氧化物酶体增殖物激活受体γ(peroxisome proliferator-activated receptor gamma,PPARγ)含有RING指状结构域,可促进p65的K48位点泛素化和降解,并且还可影响p65的K28位点的泛素化修饰[25]。含有三联体结构域的22号蛋白(tripartite motif containing 22,TRIM22)可与IKKγ形成复合物,促进K63位点泛素化,导致IKKα/β和IκBα磷酸化[26]。肿瘤坏死因子受体相关因子6(TNF receptor associated factor 6,TRAF6)可通过催化合成K63位点的多聚泛素链,使IKK激酶发生泛素化,进而激活NF-κB信号通路[27]。KCMF1作为E3泛素连接酶,可以通过K63位连接对靶蛋白进行泛素化修饰[13],但KCMF1对NF-κB信号通路分子的泛素化修饰目前仍不清楚,还需进一步探索。
综上所述,KCMF1在人CRC组织中异常高表达,敲降KCMF1可下调人CRC细胞的NF-κB信号通路的活化,并抑制CRC细胞增殖,促进细胞凋亡。
利益冲突声明:所有作者均声明不存在利益冲突。
作者贡献声明:
吴志柏:完成实验,实验分析,文章撰写;
许桂琴:实验设计和监督,指导文章修改;
张力,杨兆娟,刘昀:指导实验;
焦琨,陈泽宏,许晨:生信和数据分析;
左佑,郑宁倩,叶志谦:参与实验;
刘永忠:设计研究思路及方案,审核文章。
[参考文献]
[1] SUNG H, FERLAY J, SIEGEL R L, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA Cancer J Clin, 2021, 71(3): 209-249.
[2] GUINNEY J, DIENSTMANN R, WANG X, et al. The consensus molecular subtypes of colorectal cancer[J]. Nat Med, 2015, 21(11): 1350-1356.
[3] LAURENT-PUIG P, AGOSTINI J, MALEY K. Colorectal oncogenesis[J]. Bull Cancer, 2010, 97(11): 1311-1321.
[4] TAIXIANG W, MUNRO A J, GUANJIAN L. Chinese medical herbs for chemotherapy side effects in colorectal cancer patients[J]. Cochrane Database Syst Rev, 2005, 2005(1): CD004540.
[5] RONNEKLEIV-KELLY S M, KENNEDY G D. Management of stage Ⅳ rectal cancer: palliative options[J]. World J Gastroenterol, 2011, 17(7): 835-847.
[6] XIE Y H, CHEN Y X, FANG J Y. Comprehensive review of targeted therapy for colorectal cancer[J]. Signal Transduct Target Ther, 2020, 5(1): 22.
[7] GIANNAKIS M, MU X J, SHUKLA S A, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma[J]. Cell Rep, 2016, 17(4): 1206.
[8] HERSHKO A, CIECHANOVER A. The ubiquitin system[J]. Annu Rev Biochem, 1998, 67: 425-479.
[9] SWATEK K N, KOMANDER D. Ubiquitin modifications[J]. Cell Res, 2016, 26(4): 399-422.
[10] QI J F, RONAI Z A. Dysregulation of ubiquitin ligases in cancer[J]. Drug Resist Updat, 2015, 23: 1-11.
[11] BEILKE S, OSWALD F, GENZE F, et al. The zinc-finger protein KCMF1 is overexpressed during pancreatic cancer development and downregulation of KCMF1 inhibits pancreatic cancer development in mice[J]. Oncogene, 2010, 29(28): 4058-4067.
[12] HONG J H, KAUSTOV L, COYAUD E, et al. KCMF1 (potassium channel modulatory factor 1) Links RAD6 to UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4) and lysosomemediated degradation[J]. Mol Cell Proteomics, 2015, 14(3): 674-685.
[13] HEO A J, KIM S B, JI C H, et al. The N-terminal cysteine is a dual sensor of oxygen and oxidative stress[J]. Proc Natl Acad Sci U S A, 2021, 118(50): e2107993118.
[14] CERVIA L D, SHIBUE T, BORAH A A, et al. A ubiquitination cascade regulating the integrated stress response and survival in carcinomas[J]. Cancer Discov, 2023, 13(3): 766-795.
[15] SINGH A, CHOUDHURY S D, SINGH P, et al. Disruption in networking of KCMF1 linked ubiquitin ligase impairs autophagy in CD8+ memory T cells of patients with renal cell carcinoma[J]. Cancer Lett, 2023, 564: 216194.
[16] JANG J H. FIGC, a novel FGF-induced ubiquitin-protein ligase in gastric cancers[J]. FEBS Lett, 2004, 578(1/2): 21-25.
[17] XI Y, XU P F. Global colorectal cancer burden in 2020 and projections to 2040[J]. Transl Oncol, 2021, 14(10): 101174.
[18] YANG Y, WANG H Y, CHEN Y K, et al. Current status of surgical treatment of rectal cancer in China[J]. Chin Med J, 2020, 133(22): 2703-2711.
[19] VARLAND S, SILVA R D, KJOSÅS I, et al. N-terminal acetylation shields proteins from degradation and promotes agedependent motility and longevity[J]. Nat Commun, 2023, 14(1): 6774.
[20] ZOU J, MI L, YU X F, et al. Interaction of 14-3-3σ with KCMF1 suppresses the proliferation and colony formation of human colon cancer stem cells[J]. World J Gastroenterol, 2013, 19(24): 3770-3780.
[21] SAKAMOTO K, MAEDA S, HIKIBA Y, et al. Constitutive NFkappaB activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth[J]. Clin Cancer Res, 2009, 15(7): 2248-2258.
[22] RAJITHA B, BELALCAZAR A, NAGARAJU G P, et al. Inhibition of NF-κB translocation by curcumin analogs induces G0/G1 arrest and downregulates thymidylate synthase in colorectal cancer[J]. Cancer Lett, 2016, 373(2): 227-233.
[23] JANI T S, DEVECCHIO J, MAZUMDAR T, et al. Inhibition of NF-kappaB signaling by quinacrine is cytotoxic to human colon carcinoma cell lines and is synergistic in combination with tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) or oxaliplatin[J]. J Biol Chem, 2010, 285(25): 19162-19172.
[24] SENFT D, QI J F, RONAI Z A. Ubiquitin ligases in oncogenic transformation and cancer therapy[J]. Nat Rev Cancer, 2018, 18(2): 69-88.
[25] HOU Y Z, MOREAU F, CHADEE K. PPARγ is an E3 ligase that induces the degradation of NFκB/p65[J]. Nat Commun, 2012, 3: 1300.
[26] JI J X, DING K K, LUO T, et al. TRIM22 activates NF-κB signaling in glioblastoma by accelerating the degradation of IκBα[J]. Cell Death Differ, 2021, 28(1): 367-381.
[27] DENG L, WANG C, SPENCER E, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain[J]. Cell, 2000, 103(2): 351-361.